Section: New Results
Computational Anatomy
Geometric generative model of organ shapes: statistical properties of template shape estimation
Participants : Nina Miolane [Correspondent, Inria - Stanford] , Xavier Pennec [Inria] , Susan Holmes [Stanford] .
This work is conducted jointly with the Department of Statistics of Stanford, in the context of the associated team GeomStats and the FSCIS (France-Stanford Center for Interdisciplinary Studies) fellowship of Nina Miolane.
template, atlas, consistency, estimation theory, Expectation-Maximization algorithm, shapes, quotient space, lie group, sub-Riemannian, in-painting, neuro-geometry, visual cortex, diffusion
This work focuses on the interaction between statistics and geometry, for applications in Medical Imaging. The first part deals with a generative model of (organ) shapes and, more precisely, on the estimation of the mean shape or template. The second part of this work surveys and unveils the mathematical framework needed to extend Neurogeometry, used in 2D Computer Vision, to applications in 3D imaging.
-
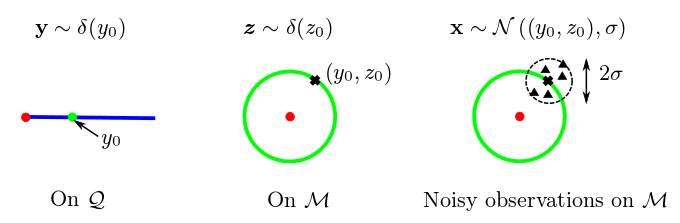
In the first part, we define a geometric statistical framework of an organ shapes generative model (see Figure 7 ). This is done through the differential geometry of quotient spaces.
-
Then, we interpret the computation of the mean organ shape (or template) through the max-max algorithm, as an approximate maximum likelihood estimation in this framework.
-
Finally, we study the statistical properties of the template computed with this procedure. More precisely, we show that the estimation is inconsistent and that the inconsistency cannot be neglected when the real template is close to the singularity of the quotient space at the scale of the ambient noise on the images [44] .
-
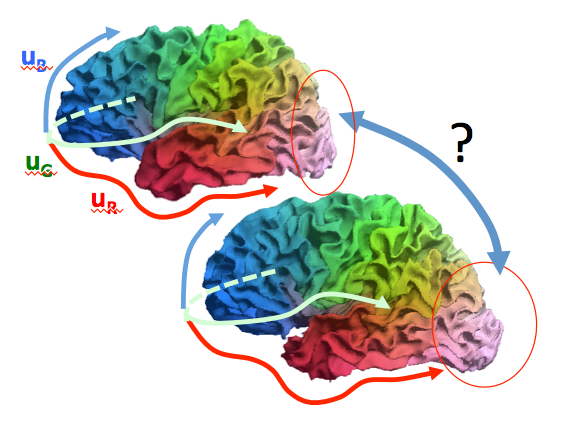
In the second part, the particularities of a 3D neurogeometry are highlighted with respect to the 2D case. They rely on the fact that 2D neurogeometry is inspired by the primary visual cortex, which codes for our 2D visual field (our retina is 2D). Imagining a 3D visual field or a 3D retina would give rise to a 3D neurogeometry.
-
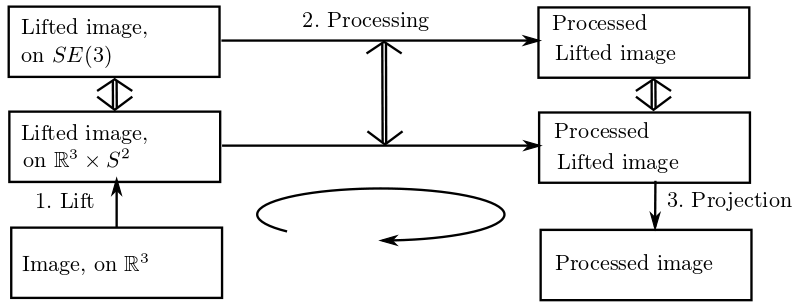
The conceptual framework of a 3D neurogeometry is more subtle, and a new level of mathematical structures arises (see Figure 8 ). Thus, inpainting (sub-Riemannian diffusion) have to be generalized.
-
Applications for in-painting or super-resolution in 3D medical images are described [43] .
Compact representation of longitudinal deformations
Participants : Raphaël Sivera [Correspondent] , Hervé Delingette, Nicholas Ayache.
This work is supported by a PhD fellowship from the University Nice Sophia Antipolis and by the European Research Council through the ERC Advanced Grant MedYMA 2011-291080 (on Biophysical Modeling and Analysis of Dynamic Medical Images).
Longitudinal modeling, Learning in manifolds, Structured sparsity.
The analysis of dynamic or longitudinal series of medical images is important to better understand the observed evolutions of the organs but also to provide robust computer aided diagnosis tools. This analysis can be performed through a reduced representation of geometric transformations capturing the deformation between 2 time points.
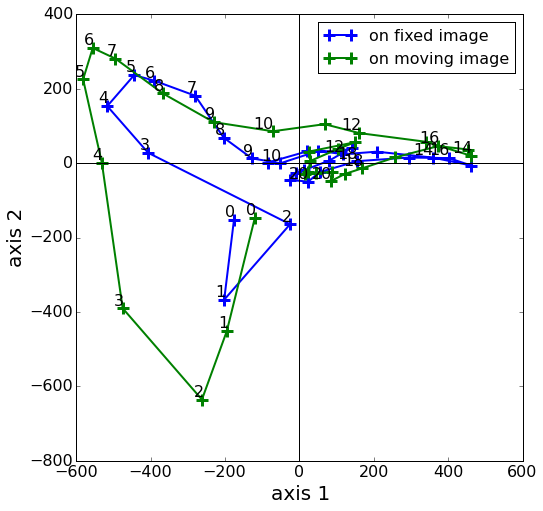
In the context of cardiac motion analysis, we proposed a framework to represent arbitrary diffeomorphisms described as Stationary Velocity Fields (SVF) in a low dimensional linear space (see fig. 9 ).
To this end, we first improved the Inverse Scaling and Squaring (ISS) algorithm from [83] to transform displacement fields into SVFs. Second, through a structured sparse decomposition of these deformations over the cardiac cycle, we provided a preliminary approach for comparing trajectories of cine-MR images between two patients.
|
Statistical analysis of heart motion
Participants : Marc-Michel Rohé [Correspondent] , Nicolas Duchateau, Maxime Sermesant, Xavier Pennec.
This work is partly supported by the FP7 European project MD-Paedigree and by the ERC Advanced Grant MedYMA 2011-291080 (on Biophysical Modeling and Analysis of Dynamic Medical Images).
Statistical analysis, Registration, Reduced order models, Machine learning
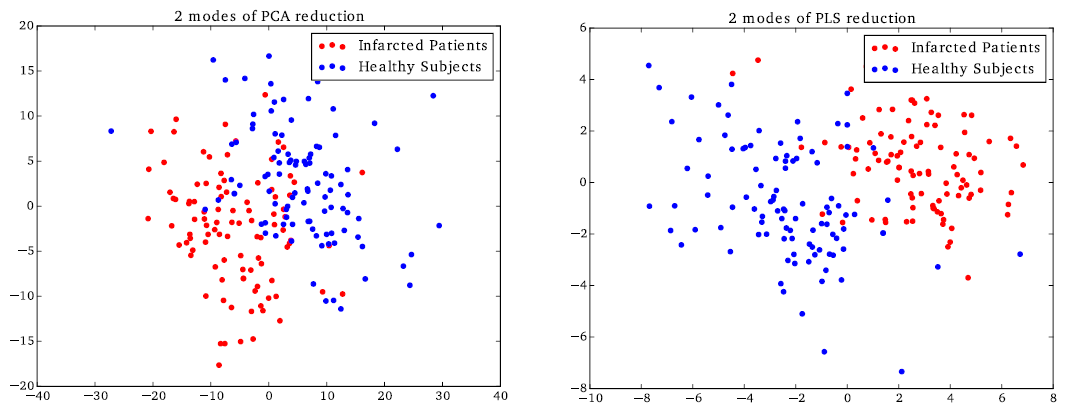
This work aims at developing statistical tools to analyze cardiac motion. In particular, we are interested in approximating complex motion models with few parameters or modes that are clinically relevant (reduced models). To this end, we have introduced a polyaffine cardiac motion model that reduces the deformation parameters to a few interpretable parameters, and the most important modes to represent the variability seen in a population are automatically selected. We then performed a group-wise statistical analysis, which relates the model parameters to clinical indices specific to a given pathology. This method was used to classify a population of healthy/infarcted hearts [48] (see Fig. 10 ), as well as to study cardiac motion of adolescents with cardiomyopathies within the European project "MD-Paedigree".
|
Statistical Learning via Synthesis of Medical Images
Participants : Hervé Lombaert [Correspondent] , Héloise Bleton, Hervé Delingette, Nicholas Ayache, Antonio Criminisi.
This work is partly supported by a grant from Microsoft Research-Inria Joint Centre and by the ERC Advanced Grant MedYMA 2011-291080 (on Biophysical Modeling and Analysis of Dynamic Medical Images).
statistical learning, synthesis
Machine learning approaches typically require large training datasets in order to capture as much variability as possible. Application of conventional learning methods on medical images is difficult due to the large variability that exists among patients, pathologies and image acquisitions. The project aims at exploring how realistic image synthesis could be used to improve existing machine learning methods.
We tackled the problem of better exploiting existing training sets, via a smart modeling of the image space, and applying conventional random forests using guided bagging [99] . Synthesis of complex data, such as cardiac diffusion images (DTI), was also done, with a refined version of [98] .
|
Then, we tackled the problem of exploiting Geometry in Data, via intrinsic representations of shapes and data [27] . Spectral decomposition (Fig. 11 ) of shapes provides a new intrinsic framework for synthesizing complex shapes such as cerebral surfaces [35] , and describing functions efficiently on these complex surfaces. This framework establishes the basics for machine learning of surface data [36] . An early application was conducted on retinotopy [57] (the study of functions in the visual cortex).
Consistency of the estimation of the template in quotient spaces
Participants : Loïc Devilliers [Correspondent] , Stéphanie Allassonnière [Ecole Polytechnique] , Xavier Pennec.
Template estimation, Fréchet mean, quotient spaces
In [24] , we studied the estimation of the template (the mean shape of our data) when the data is transformed by unknown group elements. In the case of a finite group acting isometrically on a linear space, we proved that the estimation of the template using the Fréchet mean in the quotient space is not always consistent.